Research Overview
Cells encode the instructions for life within their genomes. But this DNA is under constant assault from a variety of sources. Going outside without sunblock, drinking alcohol, smoking, and exposure to radiation are particularly serious stresses. But even just copying the genome can cause multiple forms of DNA damage. Failure to detect and reverse DNA damage can lead to errors that affect functional sequences and perpetuate from generation to generation.
Multicellular eukaryotes have evolved a wide variety of integrated pathways to sense and repair many types of DNA damage, from bulky lesions to double strand breaks. Deficiencies in these pathways can cause cells to accumulate genomic errors that lead to human diseases, including somatic cancers and Mendelian inherited genetic disorders.
But there is a bright side to DNA damage. Genome engineering systems, from nucleases to prime editors, work by targeting DNA damage and influencing repair to achieve a desired outcome. Genome engineering has revolutionized approaches to fundamental biological discovery and begun to yield cell-based therapies and cures for debilitating genetic disorders. DNA damaging genome engineering tools represent new opportunities to study repair in human cells and intensify the urgency of studying these processes so that we can better control engineering.
Research in the Corn lab seeks to understand the intersection between human DNA repair and genome engineering tools and to develop new approaches to cure human diseases using genome editing. We also use advanced genome editing to uncover the mechanisms by which cells accomplish dramatic transformations, such as the the stimulus-dependent destruction of entire organelles. We take a multidisciplinary approach to tackle these problems that includes genome-wide functional genomics, computational modeling, in vitro biochemistry and biophysics, and mechanistic cellular biochemistry.

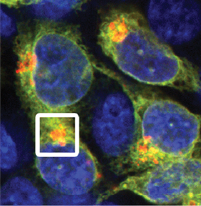 Cells keep a close eye on their contents. Misfolded proteins are marked by ubiquitin ligases and sent for destruction in the proteasome. Similarly, entire damaged organelles such as the mitochondria and endoplasmic reticulum can be marked for autophagic destruction in the lysosome. An inability to clear damaged proteins and/or organelles underlies a host of human disorders, including Alzheimer’s and Parkinson’s Disease. Organelle autophagy is also a hallmark of dramatic cellular differentiation, such as the loss of all organelles during the production of the lens or red blood cells. We are using next-generation genome engineering technologies to discover new players in the homeostasis of targets ranging from proteins to organelles. For example, using unbiased screens with complex phenotypes, we are decoding novel effectors and signaling logic that code for the destruction of each organelle. Endogenous tagging and editing approaches enable us to gain a deep understanding of the mechanisms by which these effectors operate and how they mis-fire in human disease.
Cells keep a close eye on their contents. Misfolded proteins are marked by ubiquitin ligases and sent for destruction in the proteasome. Similarly, entire damaged organelles such as the mitochondria and endoplasmic reticulum can be marked for autophagic destruction in the lysosome. An inability to clear damaged proteins and/or organelles underlies a host of human disorders, including Alzheimer’s and Parkinson’s Disease. Organelle autophagy is also a hallmark of dramatic cellular differentiation, such as the loss of all organelles during the production of the lens or red blood cells. We are using next-generation genome engineering technologies to discover new players in the homeostasis of targets ranging from proteins to organelles. For example, using unbiased screens with complex phenotypes, we are decoding novel effectors and signaling logic that code for the destruction of each organelle. Endogenous tagging and editing approaches enable us to gain a deep understanding of the mechanisms by which these effectors operate and how they mis-fire in human disease. Genome editing holds great promise to uncover the root causes of human diseases and even to reverse mutations that cause inherited genetic disorders. Our lab collaborates with clinicians and industry groups to translate genome editing approaches towards real world applications. We use genome editing to explore the mechanisms by which genomic variation causes or modifies disease, for example by introducing disease-associated mutations into human cells and measuring exact phenotypic outcomes. And we develop reagents aimed at curing genetic disorders via precision sequence replacement, for example reversing sickle cell disease or Fanconi Anemia mutations in human hematopoietic stem cells.
Genome editing holds great promise to uncover the root causes of human diseases and even to reverse mutations that cause inherited genetic disorders. Our lab collaborates with clinicians and industry groups to translate genome editing approaches towards real world applications. We use genome editing to explore the mechanisms by which genomic variation causes or modifies disease, for example by introducing disease-associated mutations into human cells and measuring exact phenotypic outcomes. And we develop reagents aimed at curing genetic disorders via precision sequence replacement, for example reversing sickle cell disease or Fanconi Anemia mutations in human hematopoietic stem cells.