Contact Us
Questions and/or comments about Corn Lab and its activities may be addressed to:
This post is all about establishing safety for CRISPR gene editing cures for human disease. Note that I did not say this post is about gene editing off-targets. We’ll get there, but you...
This post is all about establishing safety for CRISPR gene editing cures for human disease. Note that I did not say this post is about gene editing off-targets. We’ll get there, but you might be surprised by what I have to say.
Contrary to what some might say or write, most of us gene editors do not have our heads in the sand when it comes to safety. From a pre-clinical, discovery research point of view, the safety of a given gene editing technology is relatively meaningless. There are many dirty small molecules out there that you’d never want to put in a person but are ridiculously useful to help unravel new biology. Given that pre-clinical researchers have the luxury of doing things like complementation/re-expression experiments and isolating multiple independent clones, let’s dissociate everyone's personal research experience with CRISPR (all pre-clinical at this point) from questions of safety. Those experiences are useful and informative, but so far anecdotal and not necessarily tightly linked to the clinic.
Despite what we gene editors like to think, CRISPR safety is not a completely brand new world full of unexplored territory. While there are some important unanswered questions, there’s a lot of precedent. Not only are other gene editing technologies already in the clinic, but non-specific DNA damaging agents are actually effective chemotherapies (e.g. cisplatin, temozolomide, etoposide). In the latter case, the messiness of the DNA damage is the whole point of the therapy.
Here are a few thoughts about the safety of a theoretical CRISPR gene editing therapy, in mostly random order. I’ll preface this by saying that, while I have experience in the drug industry, I’m by no means an expert on clinical safety and defer to the real wizards.
Let’s start with a big point: as with any disease, the safety of a gene editing therapy is all about the indication. The safety profile of a treatment for a glioblastoma (very few good treatment options for a fatal disease with fast progression) will be very different than a treatment for eczema. And the safety tolerance of a glioblastoma treatment that increases progression-free survival by only two days is going to look different than one that increases overall survival by five years. So there won’t be One True Rule for gene editing safety, since most of the equation will be written by the disease rather than the therapy.
While most of the safety equation is about the disease, the treatment itself of course needs to be taken into account. At heart, gene editing reagents are DNA damaging agents, and so genotoxicity is a big concern. Does the intervention itself disrupt a tumor suppressor and lead to cancer? Does it break a key metabolic enzyme and lead to cell death? As mentioned above, there are plenty of DNA damaging agents that wreak havoc in the genome, but are tolerated because due to risk/reward and lack of a better alternative (I’m especially looking at you, temozolomide). The key to this point is function of what gets disrupted.
With CRISPR, we have the marked advantage that guide RNAs tend to hit certain places within the genome. We know how to design the on-target and are still figuring out how to predict and measure the off-target. But even with perfect methods for off-targets, we’d still need to do the functional test. Consider a “traditional” therapeutic (small molecule or biologic) - while an in vitro off-target panel based on biochemistry is valuable, it’s no substitute at all for normal-vs-tumor kill curves (as an example). And even those kill curves are no substitute for animal models. The long term, functional safety profile of a gene editing reagent is the key question, and with CRISPR I’d argue that we’re still too early in the game to know what to expect. The good news is that ZFNs so far seem pretty good, giving me a lot of hope for gene editing as a class.
You’d think that determining an exhaustive list of off-target sequences would be a critical part of any CRISPR safety profile. But in the example above, contrasting in vitro biochemical assays with organismal models, I consider lists of off-targets to be equivalent to the biochemical assay. I’m going to be deliberately controversial for a moment and posit that, for a therapeutic candidate, you shouldn’t put much weight on its list of off-target sites. As stated above, you should instead care about what those off-targets are doing, and for that you might not even need to know where the off-targets are located.
When choosing candidate therapeutics in a pre-clinical mode, lists of off-target sequences can be very useful in order to prioritize reagents. If one guide RNA hits two off-target sites and another hits two hundred, you’d probably choose the former rather than the latter. But what if one the two off-targets is p53? What if the two-hundred are all intergenic? Given the fitness advantage to oncogenic mutations, the math involved in using sequencing (even capture-based technologies) to detect very rare off-target sites is daunting. Being able to detect a 1 in a million sequence-based event sounds incredible, but what if you need to edit as many as 20 million cells for a bone marrow transplant? That’s twenty cells you might be turning cancerous but never even know it. Now we come right back around to function - you should care much more about the functional effect of your gene edit rather than a list of sequences. That list of sequences is nice for orders-of-magnitude and useful to choose candidate reagents, but it’s no substitute for function.
There are two big questions around gene editing immunogenicity: the immunogenicity of the reagent itself, and on-target immunogenicity if the edit introduces a sequence that’s novel to the patient. What happens when the reagent itself induces a long-term immune response? For therapies that require repeat dosing, this can kill a program (hence a huge amount of work put into humanizing antibodies). A therapy that causes someone to get very sick on the second dose is not much good, nor is it useful if antibodies raised to the therapy end up blocking the treatment. But what about in situ gene editing?
Most of in situ gene editing reagents are synthetic or bacterial and so one might raise antibodies against them, but the therapy itself is (ideally), one-shot-to-cure. In that case, as long as there’s not a strong naive immune response, maybe it doesn’t matter if you develop antibodies to the editing reagent? There are few answers here for CRISPR, and most work with ZFNs has been with ex vivo edits, where the immune system isn’t exposed to the editing reagent. Time will tell if this is a problem, and animal models will be key. Even more subtle, what happens when a gene edit causes re-expression of a “normal” protein that a patient has never before expressed (e.g. editing the sickle codon to turn mutant hemoglobin into wild type hemoglobin)?
The potential for a new immune response against a new “self” protein is probably related to the extent of the change - a single amino acid change (e.g. sickle cell) is probably less likely to cause problems than introducing a transgene (e.g. Sangamo’s work inserting enzymes into the albumin locus for lysosomal storage disorders and hemophilia). But once again, I’ve heard a lot of questions and worry about this problem but very few answers. In vivo experiments are desperately needed, and the closer to a human immune system the better.
As you’ve probably gathered by now, I have a healthy respect for functional characterization when it comes to safety. That’s why it’s absolutely critical that we keep moving forward and not let theoretical worries about arbitrary numbers of off-targets stifle innovation without data. These are tools that could some day help patients in desperate need and with few other options, so let the truly predictive functional data rule the day.
This post is the first in a new, ongoing series: what are big challenges for CRISPR-based technologies, what progress have we made so far, and what might we look forward to in the near future? I’...
This post is the first in a new, ongoing series: what are big challenges for CRISPR-based technologies, what progress have we made so far, and what might we look forward to in the near future? I’ll keep posting in this series on an irregular basis, so stay tuned for your favorite topic. These posts aren't meant to belittle any of the amazing advances made so far in these various sub-fields, but to look ahead to all the good things on the horizon. I’m certain these issues are front and center in the minds of people working in these fields, and this series of posts is aimed to bring casual readers up to speed with what’s going to be hot.
First up is CRISPR imaging, in which Cas proteins are used to visualize some cellular component in either fixed or live cells. This is a hugely exciting area. 3C/4C/Hi-C/XYZ-C technologies give great insight into the proximity of two loci averaged over large numbers of cells at a given time point. But what happens in each individual cell? Or in real time? We already know that location matters, but we’re just scratching the surface on what, when, how, or why.
CRISPR imaging got started when Stanley Qi and Bo Huang fused GFP to catalytically inactivated dCas9 to look at telomeres in living cells. Since then, we’ve seen similar approaches (fluorescent proteins or dyes brought to a region through Cas9) and a lot of creativity used to multiplex up to three colors. There’s a lot more out there, but I want to focus on the future...
What's the major challenge for live cell CRISPR imaging in the near future?
Most CRISPR imaging techniques have trouble with signal to noise. It is so far not possible to see a fluorescent Cas9 binding a single copy locus when there are so many Cas9 molecules floating around the nucleus. So far imaging has side-stepped signal to noise by either targeting repeat sequences (putting multiple fluorescent Cas9s in one spot) or recruiting multiple fluorophores to one Cas9. Even then, most CRISPR imaging systems rely on leaky expression from uninduced inducible promoters to keep Cas9 copy number on par with even repetitive loci. Single molecule imaging of Halo-Cas9 has been done in live cells, but again only at repeats. Even fixed cell imaging has trouble with non-repetitive loci. Sensitivity is also a problem for RCas9 imaging - this innovation allowed researchers to use Cas9 directed to specific RNAs to follow transcripts in living cells. But it was mostly explored with highly expressed (e.g. GAPDH) or highly concentrated (e.g. stress granule) RNAs. How can we track a single copy locus, or ideally multiple loci simultaneously, to see how nuclear organization changes over time?
Someone’s going to crack the sensitivity problem, allowing people to watch genomic loci in living cells in real time. Will we learn how intergenic variants alter nuclear organization to induce disease? Will we see noncoding RNAs interacting with target mRNAs during development? With applications this big, I know many people are working on the problem and I’m sure there will be some big developments soon.
A few weeks ago, Jacob wrote a blog post about his recent experience with posting pre-prints to bioRxiv. His verdict? “…preprints ar...
A few weeks ago, Jacob wrote a blog post about his recent experience with posting pre-prints to bioRxiv. His verdict? “…preprints are still an experiment rather than a resounding success.” That sounds about right to me. I’m bearish on pre-prints right now because the very word implies that the “real” product will be the one that eventually appears “in print”. Don’t get me wrong--I think posting pre-prints is a great step toward more openness in biology, and I applaud the people who post their research to pre-print servers. Pre-prints are also a nice work-around to the increasingly long time between a manuscript’s submission and its final acceptance in traditional journals; posting a pre-print allows important results to be shared more quickly. There’s a lot of room for improvement, though. With some changes, I think pre-print servers could better encourage a real conversation between a manuscript’s authors and readers. Here are some of my thoughts on how they might achieve that. I know there are several flavors of pre-print servers out there, but for this post I’m going to use bioRxiv for my examples.
Improve readability
It’s 2016, we’ve got undergraduates doing gene editing, but most scientific publications are still optimized for reading on an 8.5x11” piece of paper. Pre-prints tend to be even less readable--figures at the end of the document, with legends on a separate page. The format discourages casual browsing of pre-prints, and it ensures the pre-print will be ignored as soon as a nicely typeset version is available elsewhere. I will buy a nice dinner for anyone who can make pre-prints display like a published article viewed with eLife Lens.
Better editability
bioRxiv allows revised articles to be posted prior to publication in a journal, but I would like a format that makes it really easy for authors to improve their articles. Wikipedia is a great model for how this could work. On Wikipedia, the talk page allows readers and authors to discuss ways to improve an article. The history of edits to a page shows how an article evolves over time and can give authors credit for addressing issues raised by their peers. Maintaining good version history prevents authors from posting shoddy work, fixing it later, and claiming priority based on when the original, incomplete version of the article was posted.
Crowd-source peer review
Anyone filling in a reCAPTCHA to prove they’re not a robot could be helping improve Google Maps or digitize a book. What if pre-print servers asked users questions aimed at improving an article? Is this figure well-labeled? Does this experiment have all of the necessary controls? What statistical test is appropriate for this experiment? With data from many readers about very specific pieces of an article, authors could see a list of what their audience wants. It looks like we need to repeat the experiments in Figure 2 with additional controls. Everybody likes the experiments in Figure 3, but they hate the way the data are presented.
Become the version of record
Okay, this one’s a definitely a stretch goal. Right now pre-prints get superseded by the “print” version of the article, but that doesn’t need to be the case. Let’s imagine a rosy future in which articles on bioRxiv are kept completely up-to-date. Articles are typeset through Lens, making them more readable than a journal’s PDF. There’s a thriving “talk” page where readers can post comments or criticisms. Maybe the authors do a new experiment to address readers’ comments, and it’s far easier to update the bioRxiv article than to change the journal version. At that point, bioRxiv would become the best place to browse the latest research or make a deep dive into the literature. Traditional journals could still post their own versions of articles, provided they properly cite the original work, of course.
I'm going to take a step away from CRISPR for a moment and instead discuss preprints in biology. Physicists, mathematicians, and astronomers have been posting manuscripts online before peer-revie...
I'm going to take a step away from CRISPR for a moment and instead discuss preprints in biology. Physicists, mathematicians, and astronomers have been posting manuscripts online before peer-reviewed publication for quite a while on arxiv.org. Biologists have recently gotten in on the act with CSHL's biorxiv.org, but there are others such as PeerJ. At first the main posters were computational biologists, but a recent check shows manuscripts in evo-devo, gene editing, and stem cell biology. The preprint crowd has been quite active lately, with a meeting at HHMI and a l33t-speak hashtag #pr33ps on twitter.
I recently experimented with preprints by posting two of my lab's papers on biorxiv: non-homologous oligos subvert DNA repair to increase knockout events in challenging contexts, and using the Cas9 RNP for cheap and rapid sequence replacement in human hematopoietic stem cells. Why did I do this, and how did it go?
There's been some divisive opinions around whether or not preprints are a good thing. Do they establish fair precedence for a piece of work and get valuable information into the community faster than slow-as-molasses peer review? Or do they confuse the literature and encourage speed over solid science?
In thinking about this, I've tried to divorce the issue of preprints from that of for-profit scientific publication. I found that doing so clarified the issue a lot in my mind.
Why try posting a preprint? Because it represents the way I want science to look. While a group leader in industry, I was comfortable with relative secrecy. We published a lot, but there were also things that my group did not immediately share because our focus was on making therapies for patients. But in academia, sharing and advancing human knowledge are fundamental to the whole endeavor. Secrecy, precedence, and so on are just career-oriented externalities bolted on basic science. I posted to biorxiv because I hoped that lots of people would read the work, comment on it, and we could have an interesting discussion. In some ways, I was hoping that the experience would mirror what I enjoy most about scientific meetings - presenting unpublished data and then having long, stimulating conversations about it. Perhaps that's a good analogy - preprints could democratize unpublished data sharing at meetings, so that everyone in the world gets to participate and not just a few people in-the-know.
How well did it go? As of today the PDF of one paper has been downloaded about 230 times (I'm not counting abstract views), while the other was downloaded about 630 times. That's nice - hundreds of people read the manuscripts before they were even published! But only one preprint has garnered a comment, and that one was not particularly useful: "A++++, would read again." Even the twitter postings about each article were mostly 'bots or colleagues just pointing to the preprint. I appreciate the kind words and attention, but where is the stimulating discussion? I've presented the same unpublished work at several meetings, and each time it led to some great questions, after-talk conversations, and has sparked a few nice collaborations. All of this discussion at meetings has led to additional experiments that strengthened the work and improved the versions we submitted to journals. But so far biorxiv seems to mostly be a platform for consumption rather than a place for two-way information flow.
Where does that leave my thoughts on preprints? I still love the idea of preprints as a mechanism for open sharing of unpublished data. But how can we build a community that not only reads preprints but also talks about them? Will I post more preprints on biorxiv? Maybe I'll try again, but preprints are still an experiment rather than a resounding success.
PS - Most journals openly state that preprints do not conflict with eventual submission to a journal, but Cell Press has said that they consider preprints on a case-by-case basis. This has led to some avid preprinters declaring war against Cell Press' "draconian" policies, assuming that the journals are out to kill preprints for profit motives alone. By contrast, I spoke at some length with a senior Cell Press editor about preprints in biology and had an incredibly stimulating phone call - the editor had thought about the issues around preprinting in great depth, probably even more thoroughly than the avid preprinters. I eventually submitted one of the preprinted works to a Cell Press journal without issue. Though I eventually moved the manuscript to another journal, that decision had nothing to do with the work having been preprinted.
In early February the Worldwide Threat Assessment listed CRISPR as a READ MORE
In early February the Worldwide Threat Assessment listed CRISPR as a dual-use technology, one that could be used for either good or bad (think nuclear power). At the end of that same month, a delegation from the intelligence community asked to meet with me.
Five years ago, I would never have dreamed of typing that previous sentence. Now it's a day in the life.
I flew back from a Keystone meeting a day early (this one was my science vacation, just for fun) to meet with the group. I really didn't know what to expect from the delegation. I had visions of people dressed in severe suits who wore dark glasses indoors and could read my email at the press of a button. Instead, I was treated to a lively, well-rounded group of scientists, ethicists, and economists. I could have imagined any one of them walking the halls of UC Berkeley as faculty. Though one's business card was redacted in thick black sharpie (no joke).
We were joined by Berkeley's Director of Federal Relations and had an outstanding discussion lasting a few hours. I learned a lot from the group about how government educates itself and came away very impressed with the people the U.S. government charges with looking in to emerging technologies.
But I do have a point to make in bringing up this unusual visit.
As scientists, I think we should work responsibly with our new gene editing capabilities and be honest about the potential dangers. The whole point of next-gen gene editing is that it's fast, cheap, and easy. I have undergraduates volunteering in my lab who edit their first genes within a month of joining. That's exciting, but can also be scary. I agree with the threat assessment that CRISPR could be used for bad things, since it's just a tool. In fact, it might even be possible to accidentally use CRISPR in a bad way. Think AAV editing experiments designed for mice that accidentally also target human sequences.
But like I said, gene editing is just a tool. A hammer can build a house, but it can also hit someone on the head. Likewise, gaining one tool doesn't make everything easy. Try building a house with only a hammer.
Just because we now have democratized gene editing doesn't mean that bad things will start popping up left and right. Bacterial engineering has been around for a long time, but it's still hard to do bad things in that arena. There are many other barriers and bottlenecks in the way, and the same is true for bad guys who might try gene editing.
So what should we do? As gene editors, I think we should closely and enthusiastically engage with appropriate agencies. This includes federal and state bodies, and even local groups like campus EH&S. We should also be instilling a culture of responsibility and safety in the lab, even above and beyond normal safety. It's one thing for a postdoc to remember their PPE, but it's another thing to think to ask, "Should I talk to someone before I do this experiment?" Security through obscurity is not the way, but sometimes it really is better to first talk things through in a very wide forum. Remember the outcry about the H5N1 flu papers...
The idea is not to scare people. The technology isn't scary, and gene editing really isn't new. It's just easier and cheaper now, which changes the equation a bit. We should be open about risks and proactive about managing them, otherwise they'll be managed for us.
Apologies for the long delay between posts. I was teaching this last semester and also trying to get three papers out the door.
By now I’m sure most people who might read this blog have read about the clinical trial tragedy in France, READ MORE
By now I’m sure most people who might read this blog have read about the clinical trial tragedy in France, in which several people were seriously injured (one left brain dead) during a FAAH inhibitor dose escalation study. To the best of my knowledge, this is the worst such event during a clinical trial since the cytokine storms associated with TGN1412. And highly apropos to gene editing, let’s not forget Jesse Gelsinger.
I bring up this horror because genome editing is still new but racing towards the clinic. Anyone motivated to help patients with new therapies, whether based on genome editing or otherwise, should closely examine clinical failures to learn as many lessons as possible. I’m sure a detailed analysis of the trial will be forthcoming, but what could gene editors learn from an disastrous enzyme inhibitor neuro trial?
Foremost, I think we need to think hard about safety testing for in vivo gene editing. There are currently several unbiased methods to assess off-target editing events, and some of these methods look pretty good as labs pit them against one another. If working in an ex vivo setting where one can sequence modified cells, one could certainly assure oneself that a reagent is clean. And an early safety study could test how edited cells that do get cleared from the body are tolerated.
But what about therapies that would modify a gene in situ? Differential dosing of an editing reagent might affect more or fewer cells. But it could also induce more or fewer off-targets. And unlike a traditional therapeutic, after editing has happened that patient can’t go off treatment. Instead, any negative effects become part of an individual’s genetic makeup and would need to be treated like a brand-new genetic disease.
These kinds of fundamental questions make me somewhat nervous when I hear groups declare that they’ll take Cas9 into the clinic by 2017. Therapeutic discovery takes courage, since there are real people hoping for cures at the end of the road. Those patients should be both the motivator and brakes for new therapies – how can we help them as quickly as possible while simultaneously taking care that we don’t make things worse?
I want to get there as rapidly as anyone, but while the first group to take Cas9 into the clinic will no doubt be famous in the biomedical community, the first seriously negative event (god forbid) would be infamous in a much broader sense. We need to give dots all the i’s and cross all the t’s along the way.
A few hours after I wrote this, I started to wonder if I come across as a Chicken Little on this blog. That’s probably a bit funny to those in my lab, since I’m usually relentlessly optimistic about gene editing and encourage them to be boldly entrepreneurial in their research. Maybe there’s something about the internet that brings out my opposite side.
File this one under "crotchety grumblings." I started this post as a wish list for the future of CRISPR/Cas9, but came to the realization that it could apply to just about any sexy field. So instea...
File this one under "crotchety grumblings." I started this post as a wish list for the future of CRISPR/Cas9, but came to the realization that it could apply to just about any sexy field. So instead, this is a wish list for biology in general. These points are related to the dichotomy between the business of science and the pursuit of knowledge. I'm sure some of these points have come up in previous posts, but I want to get this off my chest in one place. I understand that the business of science has very logically led us to a point where the items below are natural and common, but it doesn't make them right. I'm also sure that most of these have been discussed in one form or another elsewhere, but I'm adding my $0.02. Consider this a reminder for students and postdocs who might be reading the blog.
At heart, all of the below come down to one thing. As scientists, we are trying to discover new things about the world. One assumption to most of our work is that there is an external, objective truth for us to observe. We see that truth through a glass darkly, and hence we attempt to put the external world in a form that can be understood from human context. We can do no else. To purposefully distort that truth does real harm and sets everyone back, but has unfortunately become de rigueur in some contexts.
It's a beautifully sunny day on the weekend, so I'll stop here before I dip into too much of a funk. But I'll end by saying that these are things we can fix. There is no Science with a capital S. We are scientists who are building science as we go along! We are a global community - we write our own papers, referee our own papers, write our own blog posts, form our own societies, and organize our own conferences. It's all up to us.
It seems like every time you turn around, there's another paper that mentions CRISPR/Cas9. Just how fast has the field exploded? There's nothing like data to find out! In the histogram below, each...
It seems like every time you turn around, there's another paper that mentions CRISPR/Cas9. Just how fast has the field exploded? There's nothing like data to find out! In the histogram below, each bar on the X-axis is a week (arbitrarily starting in late 2011) and the Y-axis is the number of papers in a Pubmed search for "CRISPR AND Cas9" (side note: one needs to include CRISPR in the search because "Cas9" spuriously includes results from a few labs that like to call caspases "Cas", e.g. caspase 9 = Cas9, caspase 3 = Cas3).
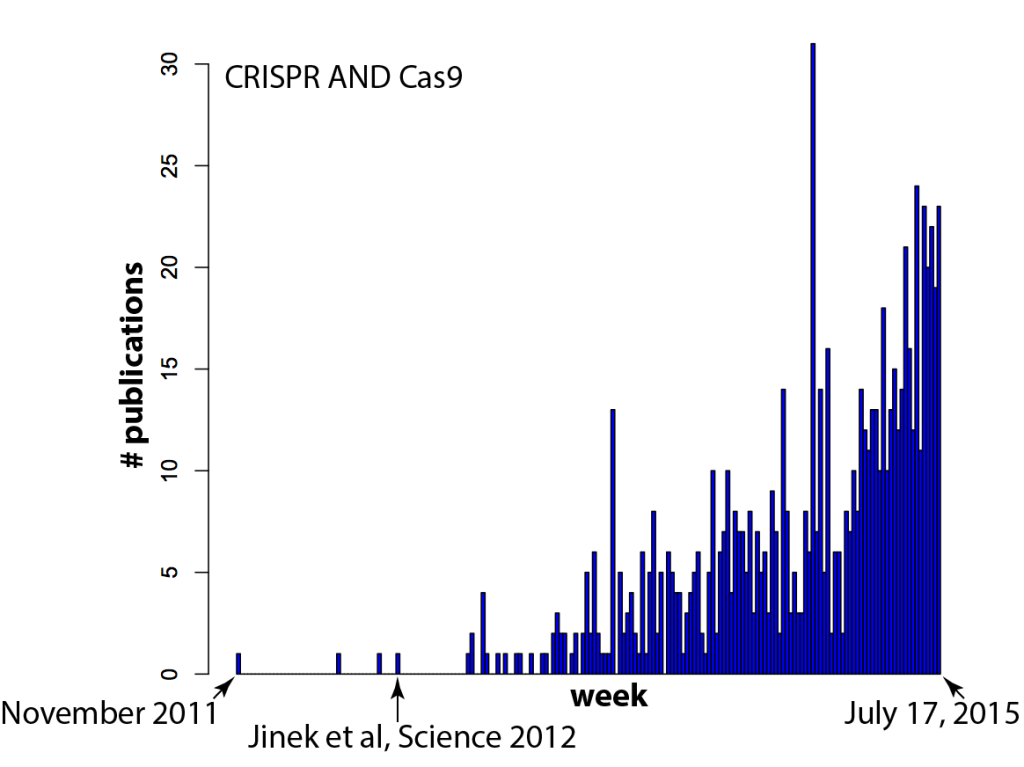
Your hunch was correct -- there IS a new paper mentioning CRISPR/Cas9 every time you turn around: several per day, in fact.
And I think (hope) that means we're reaching a tipping point. It's currently in vogue to sprinkle "Cas9" throughout a paper, even if the system is used as tool for biology and not itself the focus of the work, because it gets the attention of editors. But as more and more groups use Cas9, gene editing will become commonplace. Routine, even. And that's exactly what should happen! Can you imagine a paper these days that crows about how they used gel electrophoresis to separate proteins, or PCR and restriction enzymes to clone a gene? It's the natural course of things for disruptive technologies to quickly become just another part of the toolbox. RNAi for example, which completely upended biology not too long ago and is now used routinely and without fuss by most cell biology labs. Compare the plot above with the equivalent for RNAi (the search here was for "RNA interference" due to complications in searching "RNAi", since that also finds other hits on a viral RNA called "RNA-one". Also note that there are some false positives before Andy Fire's 1998 Nature paper).
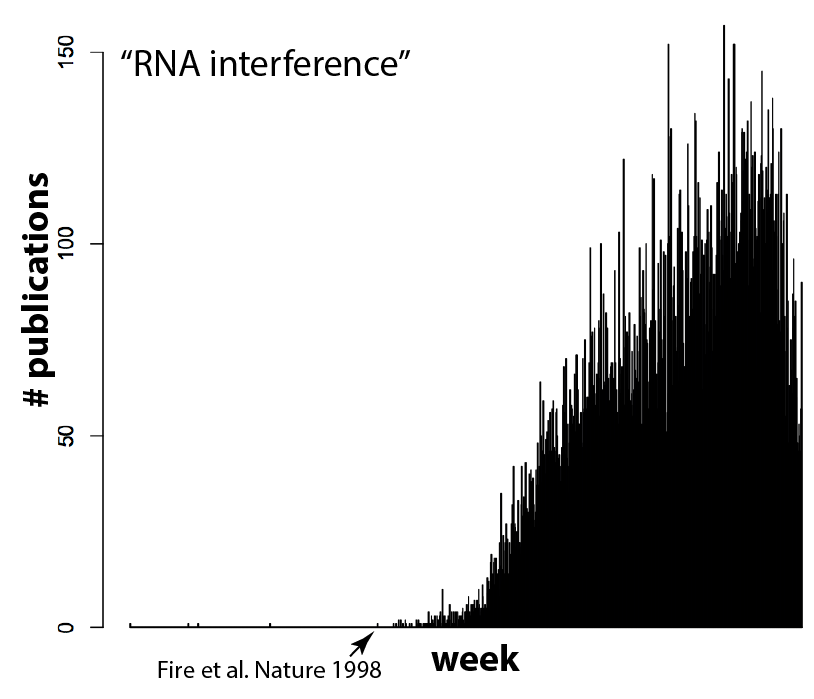
I'm looking forward to the day that CRISPR/Cas9 becomes as common-place as RNAi, since it will mean we've arrived at a new another era of biology. Want to know what that conserved genetic element does? Just remove it. Want to find out what that conserved residue does in your favorite protein? Mutate it in your organism of interest. No big deal!
That's going to be an incredible time. I'm in this for the destination, not the vehicle.
For those interested, here's how to generate the histogram
(download medline format records for a pubmed search)
# medlinedates.py
#!/usr/bin/env python
from Bio import Medline # requires Biopython
import datetime
import sys
fin = sys.argv[1]
with open(fin) as p:
records = Medline.parse(p)
for record in records:
d = record['DA']
d = datetime.date(int(d[:4]), int(d[4:6]), int(d[6:8]))
print d.toordinal()
----
(on the command line)
$ medlinedates.py [medline format file you downloaded] > dates.txt
----
(in R)
# plotdates.R
d<-read.table("dates.txt", header=F)
ranges<-append(c(seq(min(d$V1), max(d$V1), by = 7)), max(d$V1))
hist(d$V1, breaks=ranges, freq=TRUE, col="blue")
Questions and/or comments about Corn Lab and its activities may be addressed to: