Cas9 flaps
To date, most of my blog posts have talked about big picture things surrounding the business of science. But today it’s time for real data. Out today in Nature Biotechnology is our paper on new insights into Cas9 mechanism and their use to enhance precise sequence replacement in human cells.
TL;DR Cas9 “flaps” out part of the DNA target and you can take advantage of that to increase the frequency of HDR.
We started with a very simple question: what happens between Cas9 cutting and the appearance of genome edits? This could be answered in a variety of interesting ways, including elucidation of DNA repair mechanisms. We chose to go even simpler and figure out how Cas9 and sgRNA interact with and dissociate from a DNA target.
Our first surprise came when we found that catalytically inactive dCas9 takes a very long time to dissociate from DNA. As in, six hours or more! Far longer than one might expect from an enzyme that’s basically a retargetable restriction enzyme. Our second surprise came when we found that active and inactive Cas9 behave identically in this respect – even after Cas9 cuts a DNA target, it stays bound for six hours. This length of time was significant to us, since it made sense of a mystery. When one makes double strand breaks in eukaryotic cells using radiation, they are repaired very rapidly. But when Cas9 makes double strand breaks it instead takes (wait for it) about six hours for them to be repaired.
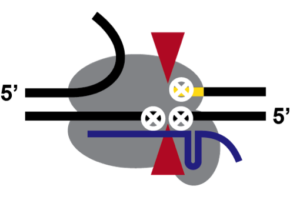
Using labeled DNA molecules, we found that Cas9 globally holds on to both sides of a cut duplex. That is, neither half duplex floats away. But we next wondered about the more fine-grained nature of the complex after cutting. That’s when we got our third surprise. It turns out that when Cas9 holds onto the four strands of the cleaved duplex (think about that one for a moment), it’s only holding on tight to three of them. The DNA strand opposite the PAM on the non-target strand (the one not bound by the sgRNA) is flapping in the breeze. So much so that in vitro we could anneal a complementary piece of ssDNA. dCas9 does the same thing, but since it doesn’t cut the target DNA there’s now an intact DNA bubble formed opposite the sgRNA. If that description is leaving you cross-eyed, here’s a picture of what I mean (gray is Cas9, blue is the sgRNA, yellow is the PAM, red are the active sites, and white x’s are the place where Cas9 is tightly holding the target).
The ability to anneal complementary single stranded oligonucleotides to a DNA flap put us in mind of recent work from Nancy Maizels’ lab, which described a nick repair structure in which single stranded repair templates annealed to one side of a flap. Turning to human cells, we asked if oligonucleotide donors capable of annealing to the Cas9 flap could be used for sequence replacement. And we were very pleasantly surprised. The Cas9 flap is on the order of 20-30 nucleotides, and indeed we found that editing donors complementary to the flap by about 30 nucleotides consistently worked the best of all donors. Absolute frequencies varied, but at one locus we reproducibly observed up to 60% sequence replacement. Making the complementary region shorter decreased efficiency as one would expect, and making it longer also caused a decrease, we think because it would need to intrude into the intact duplex outside of the flap.
Finally, we decided to try a crazy experiment. We wondered if the bubble formed by catalytically inactive dCas9 could be used to anneal a single stranded donor in cells, and if that cell could be tricked into using it for repair. By targeting three dCas9s to a precisely spaced region matching the length of a single stranded donor and all on the same strand of DNA, we did observe sequence replacement rates of a bit less than 1%. This is by no means a large number, but it was attained without any of the error-prone repair that normally accompanies Cas9 cutting. We still don’t know the mechanism underlying dCas9 editing, but it could be very useful to tackle genetic diseases in which sequence replacement holds some kind of fitness advantage but error-prone repair of the gene would be disastrous (e.g. if breaking the gene is worse than leaving the mutation alone).
I’m very curious about the unexpected flappy mechanism of Cas9 and wondering if it plays some part in Cas9’s role in CRISPR anti-viral biology. I’m also more and more intrigued in the mechanism by which single stranded donors are used by the cell. Eric Hendrickson and Nancy Maizels have some great stories on that front, and my lab has some data that seem to shed some light. On a more practical note, Cas9 is currently very good at breaking genes, but its ability to precisely introduce mutations has lagged behind. I’m optimistic that these simple approaches to designing a sequence replacement experiment we discovered, which don’t rely on chemical manipulation of cells, can be useful in tackling a variety of genetic diseases. More on that soon.
This manuscript was put together by a stellar team: postdoc Chris Richardson, technician Jordan Ray (very soon on his way to grad school), former lab manager Gemma Curie, and postdoc Mark DeWitt. Congrats to them!





I also tried to figure out how Cas9 and sgRNA interact with and dissociate from a DNA target, but at the beginning this failed and I gave up, chose to do something else. You guys are great.
Hi Jacob, interesting post and the associated papers were nice to read. I’m setting up some Crispr experiments to target a few genomic loci, and the increased repair efficiency shown in the Nature Biotech paper could greatly increase my chances of getting the right edits. There’s one thing I was wondering about: When you use the H840A Cas9 nickase (only cut adjacent to the PAM and not the other strand), do you still get the strand flapping out? If yes, this would be a great way to precisely edit difficult loci with the ssDNA donor, for which a double nickase is not possible or is too inefficient, while reducing off-target effects. What are your thoughts about this, and maybe you already tried this in the lab?
Indeed, if you nick the PAM-containing strand, then the nicked strand still flaps.