INCREASING HEMOGLOBIN HBA2 BY REPAIRING THE HBD PROMOTER, PUBLISHED IN ELIFE
Erythrocytes, or red blood cells, carry hemoglobin and circulate throughout the body to supply oxygen. β-hemoglobinopathies, such as sickle cell disease and β-thalassemia, are the most common genetic diseases worldwide and are caused by mutations affecting the structure or production of β-globin subunits in adult hemoglobin. These conditions result in anemia and organ damage, and available treatment options are limited. Stem cell transplantation is currently the only curative approach, although its feasibility relies on the availability of a suitable donor.
Hemoglobin is a tetrameric protein composed of 2 α-like (HBA) and 2 β-like subunits (HBB). Hemoglobin A1 (HbA1) constitutes 97% of adult hemoglobin, while Hemoglobin A2 (HbA2) makes up 2-3%. HbA2 is composed of two α-globin subunits and two δ-globin (HBD) subunits. HBD is a homologous to HBB gene, but with much lower expression compared to HBB due to a weak promoter. Currently, many efforts are focused on increasing fetal hemoglobin (HbF) to treat the β-hemoglobinopathies. But HbA2 is more similar to HbA1 and is already expressed at low levels in all adult red blood cells. What if we were to increase HbA2 levels? Could they potentially compensate for beta-globin deficiency? Can genome editing technologies be used to boost transcriptional activity of the endogenous HBD promoter to increase HbA2 levels? Mandy Boontanrart, a Postdoc in our lab, was eager to discover the answers to these questions.
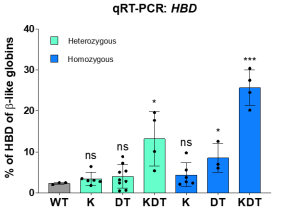
Using CRISPR-Cas9 genome editing, we inserted various transcription factor binding sequences into the endogenous HBD promoter. Team efforts yielded positive results as we successfully increased the transcriptional activity in HUDEP-2 immortalized erythroid progenitor cells, resulting in a significant upregulation of HBD expression. Despite roughly equal homology-directed repair rates between all promoter designs, we observed a significant increase in HBD only for the design with all three elements (KLF1, β-DRF, and TFIIB). We next explored whether endogenous editing of the HBD promoter can be accomplished in bone marrow stem cells. We found up to 46% HBD expression in clonal populations. We also tested a small molecule drug that enhances HDR outcomes by inhibiting the NHEJ pathway and observed an increase in the percent of HDR alleles in pooled edited bone marrow stem cells.
While our findings provide key mechanistic insight into the globin gene regulation, several questions remain to be tackled. Is heterozygous knock-in of the promoter design in β-hemoglobinopathy cells is sufficient to ameliorate disease phenotypes? What is the safety profile of this strategy?
Overall, our work is a promising approach for restoring hemoglobin levels in red blood cells. This strategy might open new therapeutic avenues for to treating beta-hemoglobinopathies in the future.
For more, check out our paper, it is now out in Elife!
Note: Excitingly, Mandy is now leading an ETH spin-off, building upon the findings of the paper, check out their brand-new website https://www.ariyabio.ch/!