CDN transporter collaboration with David Raulet published in Nature
Cells have many ways to figure out that something is wrong. One of these is cGAS, which makes a cyclic dinucleotide (CDN) to activate STING innate immune signaling....
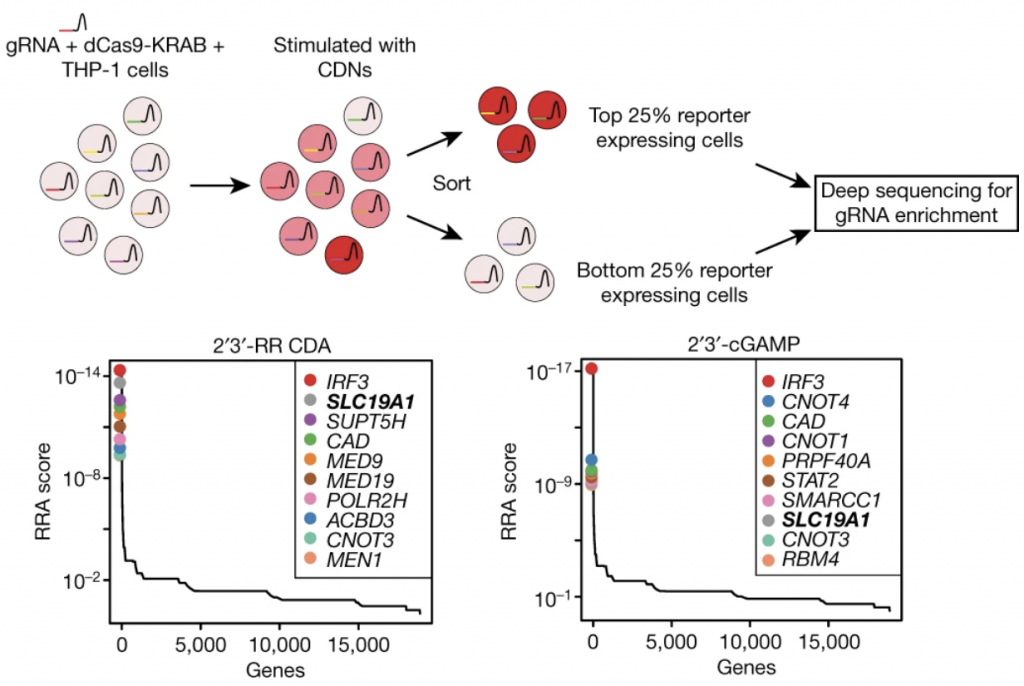
Cells have many ways to figure out that something is wrong. One of these is cGAS, which makes a cyclic dinucleotide (CDN) to activate STING innate immune signaling. CDNs are made during bacterial infection and tumor progression, and CDN derivatives are in development to re-activate immune cells next to a tumor. But how do CDNs secreted into the environment get into a target cell? This was the question asked by David Raulet’s lab, who collaborated with us on a genome-wide CRISPRi screen to find the CDN transporter. We helped the Raulet lab identify the folate transporter (SLC19A1) as a CDN importer. The Raulet lab plus further collaboration with Joshua Woodward’s lab figured out the mechanism. Lingyin Li’s lab also identified the folate transporter in a parallel collaboration with Mike Bassik, reported in Molecular Cell. Congrats to former lab members Benjamin Gowen and Stacia Wyman, who were authors on the paper, now out in Nature!

 Welcome to Zac Kontarakis, who is the head of the new Genome Engineering and Measurement Lab (GEML). The GEML is a new hub, jointly developed by Jacob Corn and...
Welcome to Zac Kontarakis, who is the head of the new Genome Engineering and Measurement Lab (GEML). The GEML is a new hub, jointly developed by Jacob Corn and...