Three timescales of impact for next-gen genome editing
This post expands on a slide that I often present in seminars: what is the scale (in time and impact) of next generation genome editing? I’m not restricting...
This post expands on a slide that I often present in seminars: what is the scale (in time and impact) of next generation genome editing? I’m not restricting this to CRISPR/Cas9, because the field is moving so fast that it’s anyone’s guess whether we’ll soon see a next next-gen (Cas10?). But the accelerator has been pressed firmly to the floor, and there’s no going back. To avoid overuse of speculative words like “might” and “could”, I’ll just speak as if I have a crystal ball. But futurism is often a fallacy and the genome editing field is only 2 years old and moving very quickly so consider what’s below a sketch at best and random guessing at worst.
Edit: Here I’m focusing on just a few areas out of many. There are very exciting things on the horizon for editing of crops and livestock, synthetic biology in normally difficult systems, and much more. I’m leaving all of that aside for now as fodder for another post.
Short: In the next few years I think we’ll see greater adoption of genome editing in many labs, both academic and industrial. This will mostly be what I call “RNAi v2.0” — disruption of genes in a very fast and easy mode (either via CRISPRcutting or CRISPRinhibition). This will extend to both human cells and model organisms, but the scope accessible for reverse genetics will be greatly expanded. Now that more and more genomes are sequenced, we’ll finally have a way to figure out what biologies underly all of those great annotations in those organisms (reverse genetics) or screen for which genes are responsible for incredible phenotypes (forward genetics). How do salamanders regenerate limbs? How do some fungi turn insects into zombies? What are the roles of genes expressed during Plasmodium infection? Does ablating gene X slow tumor progression in this model system? Are all of these genes really necessary for epithelial differentiation in the gut? These kinds of questions will be broadly answerable in both academic and industrial research settings: fundamental discoveries that will accelerate and broaden our understanding of the world around us.
Medium: Within five years true gene editing (surgically replacing one sequence with a defined replacement) will have matured and be as easy in human cells and model organisms as plasmid mutation currently is bacteria. We’re already starting to see some hints of this on the horizon, so maybe this should even be in the “short” bin. But I think a lot of current work is focused on very low hanging fruit (important though it is), and there’s still no clear path towards quickly and robustly engineering silent or deleterious variants, for example mutants with a fitness disadvantage. So this one goes into “medium term”. Surgical introduction of mutation would be huge for any number of basic biologies, since it would enable one to readily ask reductionist and mechanistic questions in the context of a living cell or organism without confounding factors. On the translational front, in the medium term gene editing will totally change the way preclinical research is carried out. Custom-designed safety models (e.g. humanized rats), highly engineered cell lines to meld target and phenotypic screening, synthetic biology for enhanced drug production, and so on. People have been wanting to do these things for a long while and they might take a little longer to achieve in industry only because the focus will include robustness of the systems rather than purely speed, but they’re coming. More relevant to the general public, in the medium term we’ll start to see the widespread clinical emergence of ex vivo therapies that take advantage of gene editing, especially in the hematopoietic system. Clinical research and trials are already ongoing here (e.g. Sangamo’s work with ZFN knockout of CCR5 for HIV), but now I’m talking about FDA approval and widespread use of an edited product as a therapeutic. The trial data has so far been very impressive on many fronts, but time will tell and the finish line is always further away than you think.
Long: Since the likelihood of anyone accurately predicting at this timescale is quite low, rather than make any specific predictions I’ll instead wax philosophic. Here we’re starting to talk about disruptive science fiction entering our lives in a real way. Things like in vivo editing in adult or postmitotic tissues. Sci-fi may actually be an apt comparison and offers a few positive examples of successful prognostication: Edward Bellamy predicted credit cards in 1888 and Arthur C. Clarke described communications satellites in 1945. And in a way, media of all kinds has been preparing us for genome editing for decades. I was recently asked how I explain what genome editing is and why it’s practically beneficial. But the thing is, I actually don’t need to do much explaining. I’ve talked about genome editing with taxi drivers, hair dressers, graphic designers, high school students, and Hollywood actresses. Everyone gets it right away. You don’t need to know a thing about Cas9 or mechanisms of DNA break repair to understand genome editing. Most people very quickly understand what genome editing is and they see how much good it could do. But everyone also sees how much harm might come if we’re reckless and how much care should be taken. So in the long term, our relationship with genetic diseases will fundamentally change. I’m not necessarily talking about germline editing, since one might have the same outcome with the ability to replace affected tissues with edited tissues. There is the opportunity for real and permanent cures for terrible diseases in which people currently just make do. That’s powerful stuff. But it’s a long road, and there’s a lot left to be done.

 Cas9 bound to sgRNA and DNA target
Cas9 bound to sgRNA and DNA target HnH domain removed (sgRNA in orange, DNA in blue)
HnH domain removed (sgRNA in orange, DNA in blue) The DNA and sgRNA can be removed from the model (PAM in green)
The DNA and sgRNA can be removed from the model (PAM in green) The DNA strands can even be separated at the actual Cas9 cleavage site
The DNA strands can even be separated at the actual Cas9 cleavage site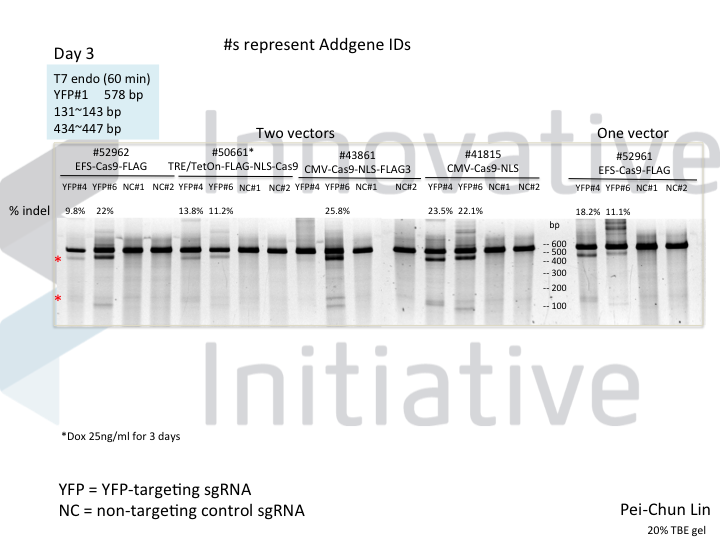
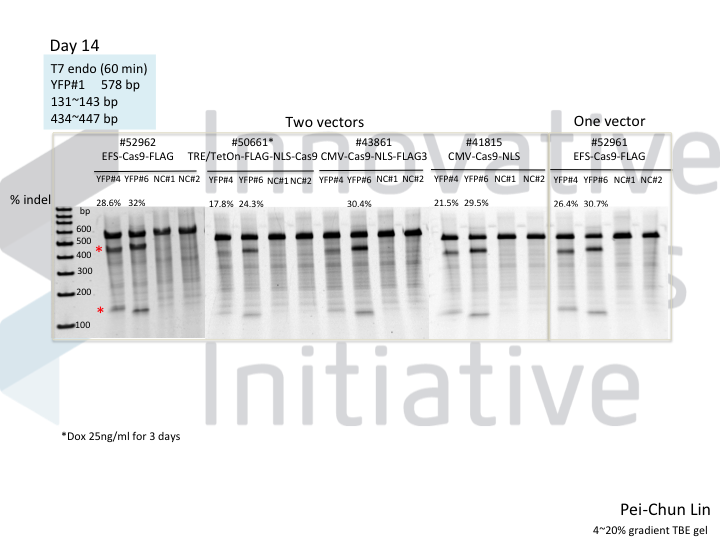
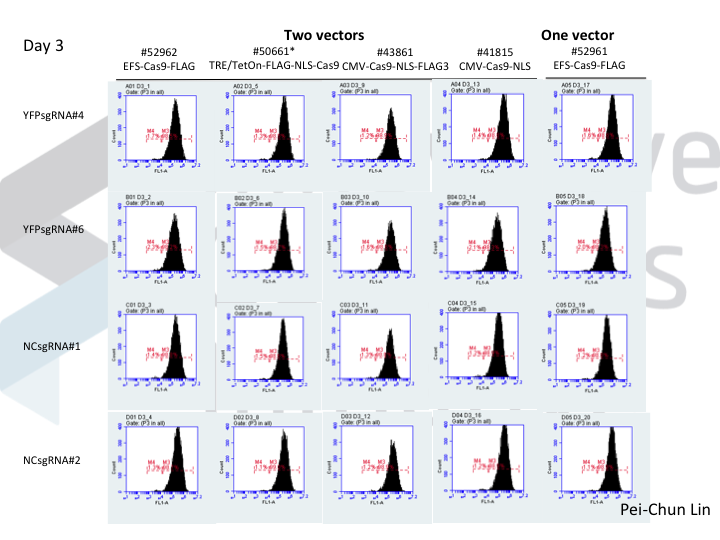
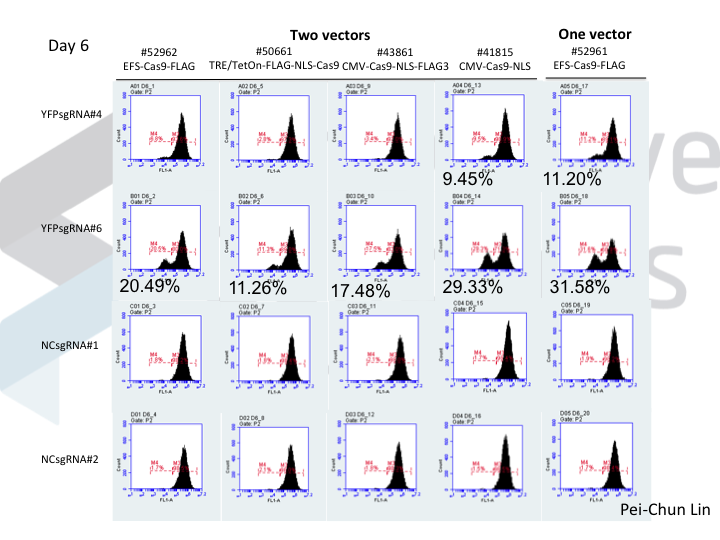


 Welcome to Lena Kobel, who joins the lab as a Cell Line Engineer. Lena has a long history in genome engineering, with previous experience in Martin Jinek’s...
Welcome to Lena Kobel, who joins the lab as a Cell Line Engineer. Lena has a long history in genome engineering, with previous experience in Martin Jinek’s...